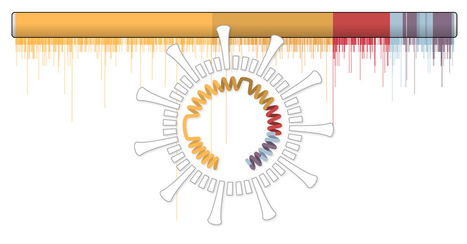
A study involving more than 5,000 COVID-19 patients in Houston finds that the virus that causes the disease is accumulating genetic mutations, one of which may have made it more contagious. According to the paper published in the peer-reviewed journal mBIO, that mutation, called D614G, is located in the spike protein that pries open our cells for viral entry. It’s the largest peer-reviewed study of SARS-CoV-2 genome sequences in one metropolitan region of the U.S. to date.
The paper shows “the virus is mutating due to a combination of neutral drift — which just means random genetic changes that don’t help or hurt the virus — and pressure from our immune systems,” said Ilya Finkelstein, associate professor of molecular biosciences at The University of Texas at Austin and co-author of the study. The study was carried out by scientists at Houston Methodist Hospital, UT Austin and elsewhere.
During the initial wave of the pandemic, 71% of the novel coronaviruses identified in patients in Houston had this mutation. When the second wave of the outbreak hit Houston during the summer, this variant had leaped to 99.9% prevalence. This mirrors a trend observed around the world. A study published in July based on more than 28,000 genome sequences found that variants carrying the D614G mutation became the globally dominant form of SARS-CoV-2 in about a month. SARS-CoV-2 is the coronavirus that causes COVID-19. So why did strains containing this mutation outcompete those that didn’t have it?
Perhaps they’re more contagious. A study of more than 25,000 genome sequences in the U.K. found that viruses with the mutation tended to transmit slightly faster than those without it and caused larger clusters of infections. Natural selection would favor strains of the virus that transmit more easily. But not all scientists are convinced. Some have suggested another explanation, called “founder’s effects.” In that scenario, the D614G mutation might have been more common in the first viruses to arrive in Europe and North America, essentially giving them a head start on other strains.



 Your new post is loading...
Your new post is loading...